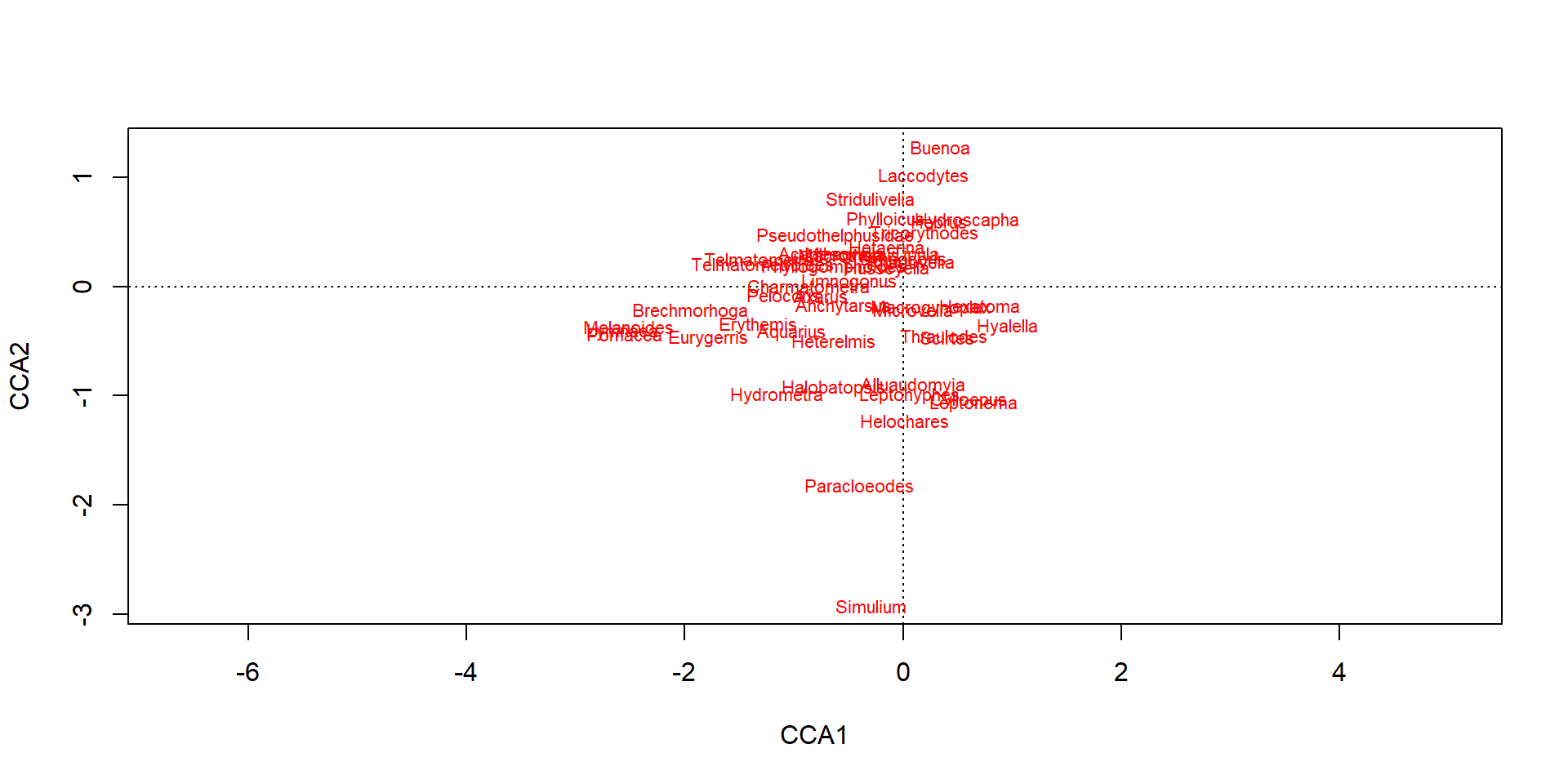
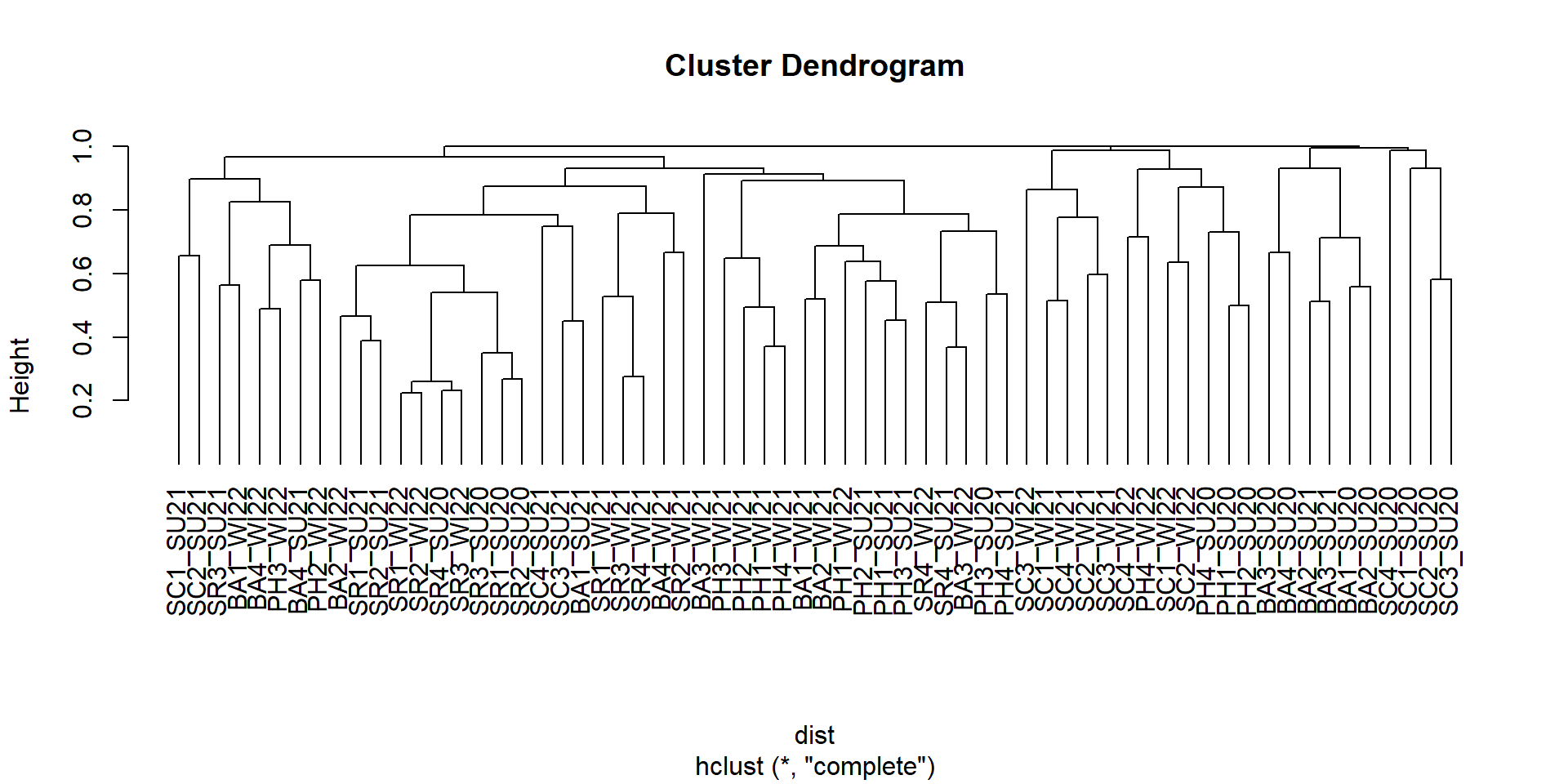
Acanthagrion Alluaudomyia Anchytarsus Aquarius Axarus Brechmorhoga
SC1_SU20 1 0 0 0 1 0
SC2_SU20 0 0 0 0 0 0
SC3_SU20 2 0 0 0 0 0
SC4_SU20 13 0 0 0 0 15
BA1_SU20 11 0 0 0 0 0
BA2_SU20 6 0 0 0 0 0
Buenoa Charmatometra Cylloepus Erythemis Eurygerris Halobatopsis
SC1_SU20 0 0 0 0 3 0
SC2_SU20 0 0 0 1 50 0
SC3_SU20 0 0 0 2 24 0
SC4_SU20 0 0 0 0 1 0
BA1_SU20 50 0 0 0 0 0
BA2_SU20 19 0 0 0 0 0
Hebrus Helochares Hetaerina Heterelmis Hexatoma Husseyella Hydrometra
SC1_SU20 0 0 0 0 0 0 0
SC2_SU20 0 0 0 0 0 0 0
SC3_SU20 0 0 0 0 0 0 0
SC4_SU20 0 0 0 0 0 0 0
BA1_SU20 0 0 1 0 0 0 0
BA2_SU20 0 0 3 0 0 0 0
Hydroscapha Laccodytes Leptohyphes Leptonema Limnocoris Limnogonus
SC1_SU20 0 0 0 0 0 0
SC2_SU20 0 0 0 0 0 0
SC3_SU20 0 0 0 0 0 0
SC4_SU20 0 0 0 0 0 0
BA1_SU20 0 46 1 1 3 0
BA2_SU20 0 70 0 1 3 0
Macrogynoplax Macrothemis Mesovelia Microvelia Paracloeodes Pelocoris
SC1_SU20 0 1 0 0 0 9
SC2_SU20 0 0 0 0 0 3
SC3_SU20 0 0 0 0 0 6
SC4_SU20 0 0 0 0 0 0
BA1_SU20 1 0 0 0 0 5
BA2_SU20 0 0 0 0 0 1
Phyllogomphoides Phylloicus Rhagovelia Scirtes Simulium Stridulivelia
SC1_SU20 0 8 0 0 0 0
SC2_SU20 0 0 0 0 0 0
SC3_SU20 0 1 0 0 0 0
SC4_SU20 0 0 0 0 0 0
BA1_SU20 4 11 32 0 0 0
BA2_SU20 0 9 0 0 0 0
Telmatometra Telmatometroides Thraulodes Tipula Tricorythodes
SC1_SU20 0 0 0 0 0
SC2_SU20 0 0 0 0 0
SC3_SU20 0 0 0 0 0
SC4_SU20 0 0 0 0 0
BA1_SU20 0 0 26 0 1
BA2_SU20 0 0 56 0 5
Melanoides Lymnaea Pomacea Pseudothelphusidae Hyalella
SC1_SU20 69 13 8 0 0
SC2_SU20 0 0 5 0 0
SC3_SU20 0 1 0 0 0
SC4_SU20 0 0 0 2 0
BA1_SU20 0 0 0 0 0
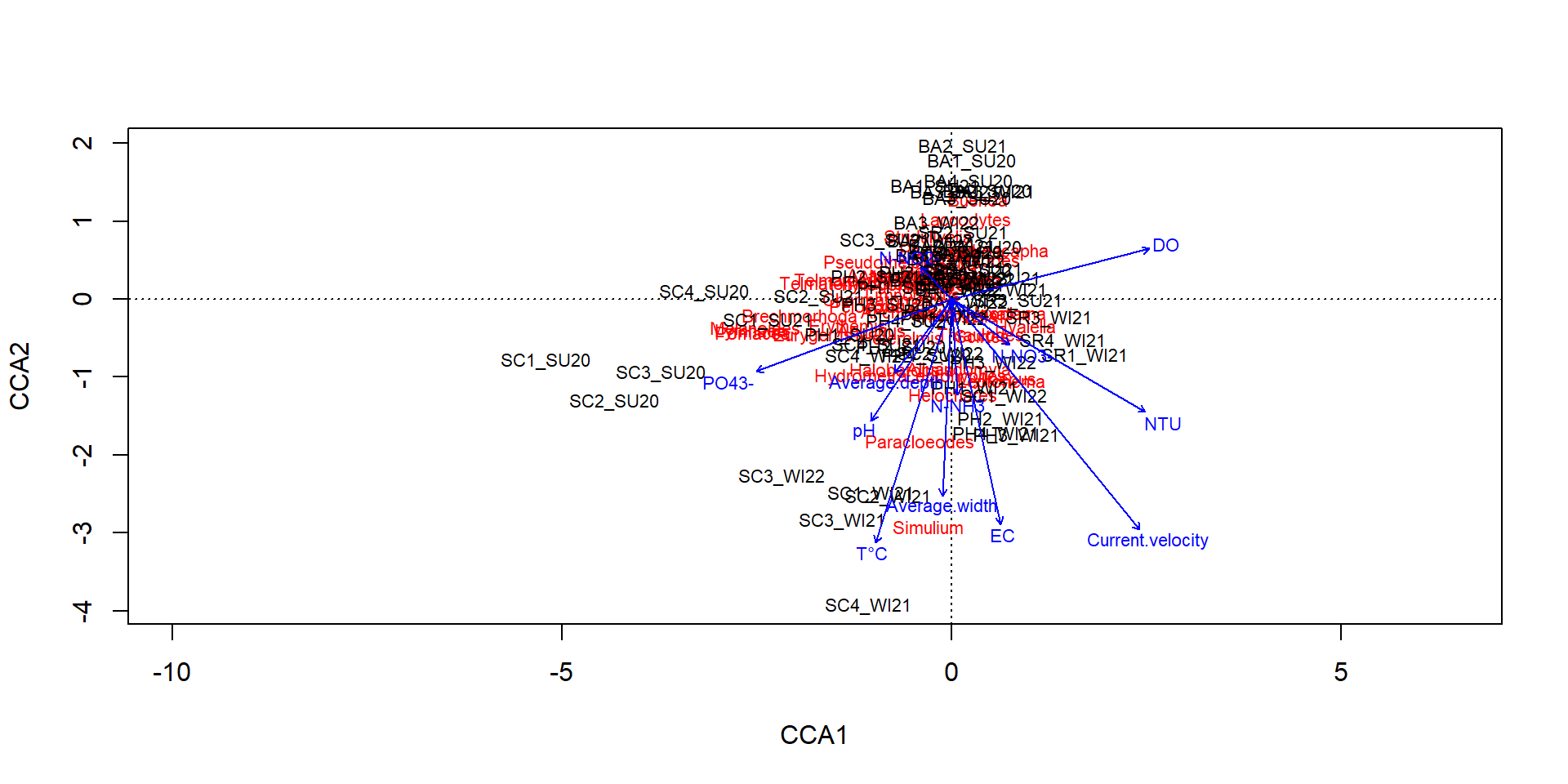
BA2_SU20 0 0 0 3 0 N-NO3- N-NH3 N-NO2- PO43- pH DO EC NTU T°C
SC1_SU20 0.02 0.09 0.003 1.20 9.165 1.840 489.0 8.10 22.180
SC2_SU20 0.02 0.10 0.003 1.12 9.230 1.860 489.0 8.55 22.495
SC3_SU20 0.02 0.13 0.005 3.86 9.115 1.905 472.0 7.70 23.035
SC4_SU20 0.02 0.18 0.003 4.09 8.745 1.985 483.5 8.15 22.445
BA1_SU20 0.05 0.03 0.006 0.73 8.200 5.950 441.5 11.40 19.300
BA2_SU20 0.04 0.10 0.005 0.72 8.940 5.950 166.5 11.85 19.945
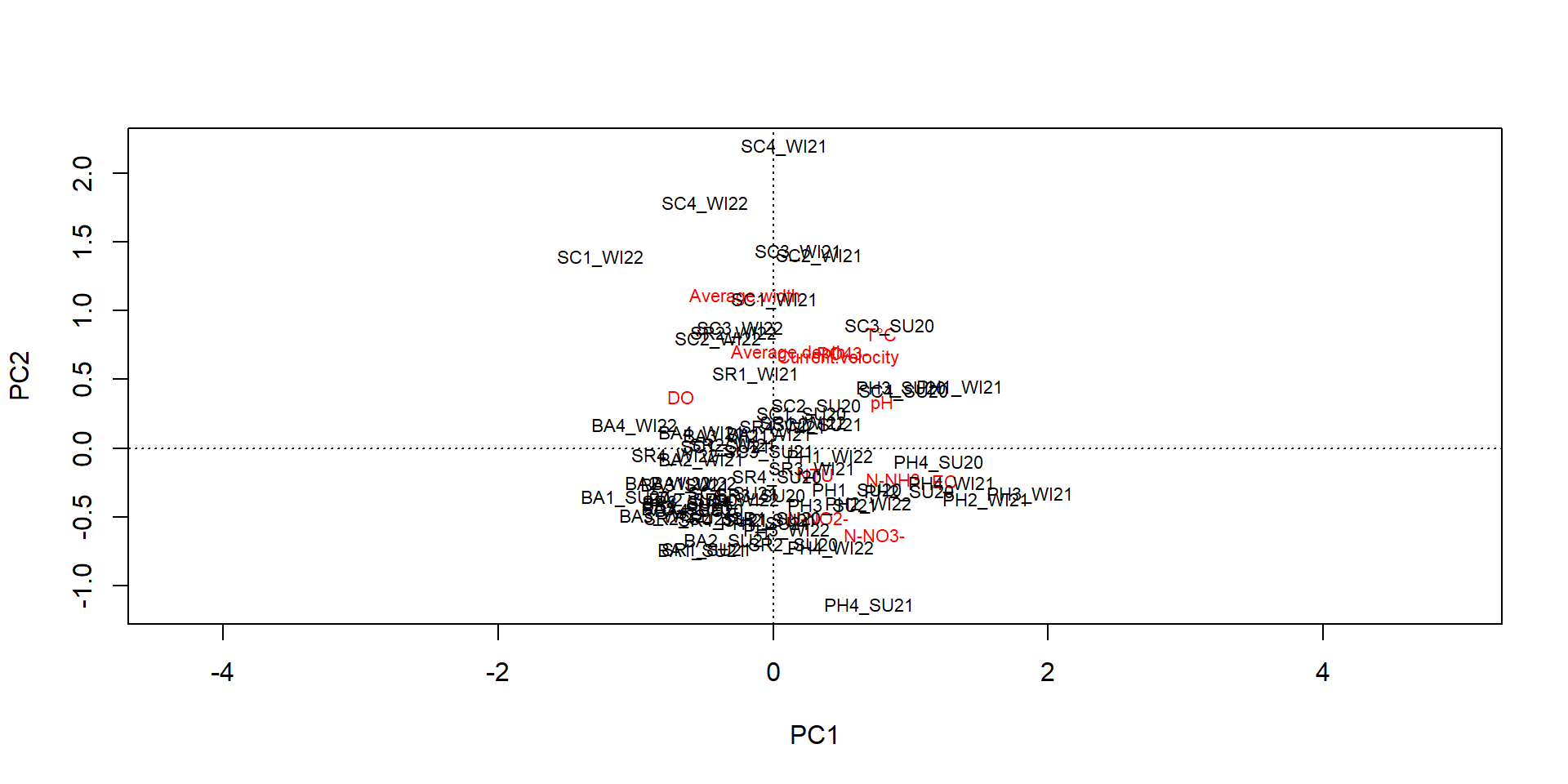
Current.velocity Average.depth Average.width
SC1_SU20 0.3454020 0.055 0.300
SC2_SU20 0.1731571 0.055 0.400
SC3_SU20 0.1848344 0.050 0.825
SC4_SU20 0.4827096 0.025 0.130
BA1_SU20 0.2882227 0.065 0.140
BA2_SU20 0.2490695 0.025 0.255