Modelos Jerárquicos de Comunidades de Especies en R
Hierarchical Models of Species Communities (HMSC)
Diego Nieto Lugilde
Universidad de Córdoba (España)
2024-10-18
Cargamos algunos datos
Preparamos un poco los datos
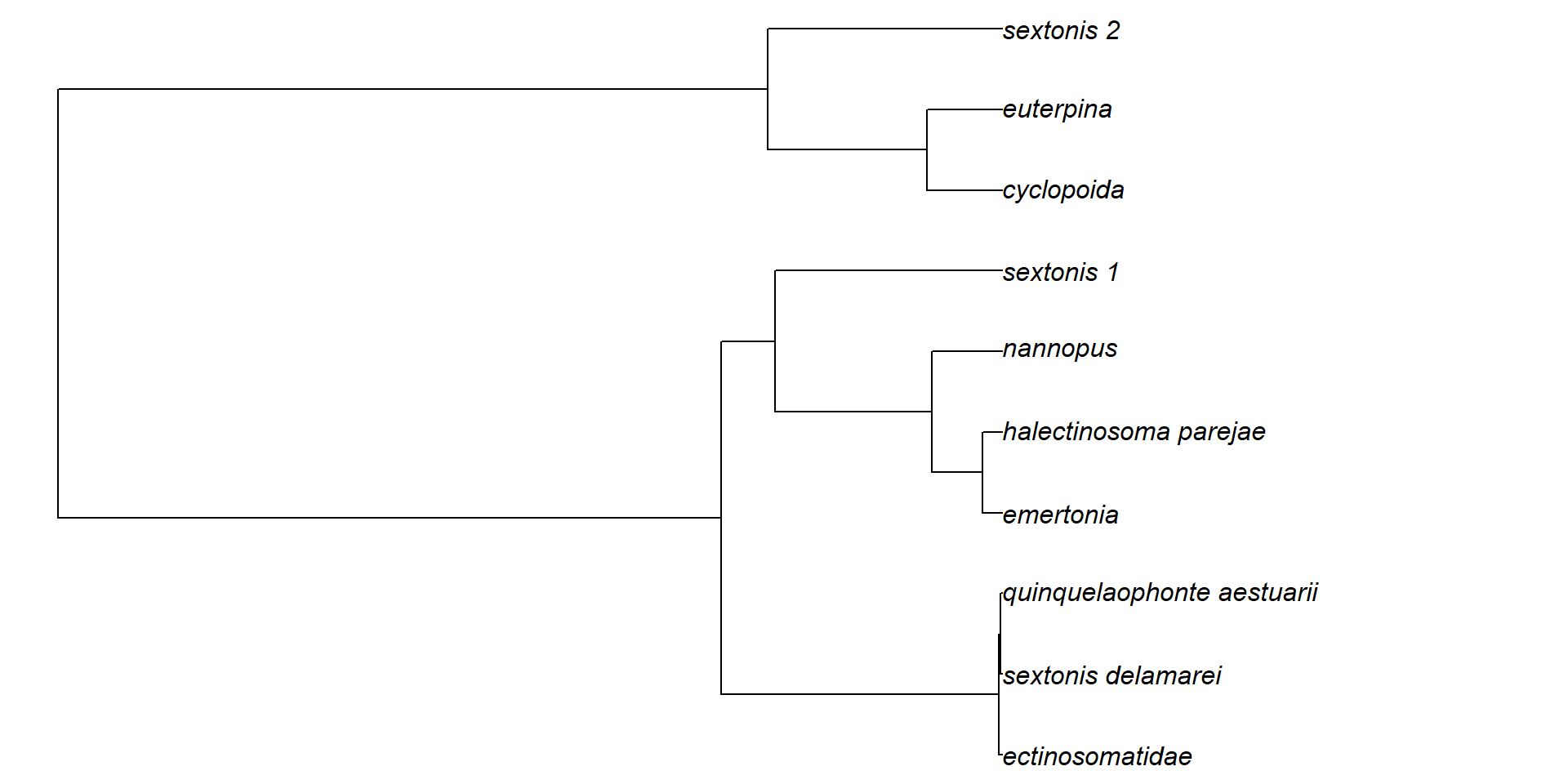
Simulamos unos datos filogenéticos
Definimos el diseño experimental
Especificamos la estructura del modelo
m0 <- Hmsc(Y = comm,
XData = envm,
XFormula = ~1,
studyDesign = studyDesign,
ranLevels = list(rio = rioL, mes = mesL),
distr = "lognormal poisson")
m1 <- Hmsc(Y = comm,
XData = envm,
XFormula = ~materia_org + poly(temperatura ,degree = 2, raw = TRUE) + salinidad + ph,
distr = "lognormal poisson")
m_env <- Hmsc(Y = comm,
XData = envm,
XFormula = ~materia_org + poly(temperatura ,degree = 2, raw = TRUE) + salinidad + ph,
studyDesign = studyDesign,
ranLevels = list(rio = rioL, mes = mesL),
distr = "lognormal poisson")
m_env_phy <- Hmsc(Y = comm,
XData = envm,
XFormula = ~materia_org + poly(temperatura ,degree = 2, raw = TRUE) + salinidad + ph,
phyloTree = phy,
studyDesign = studyDesign,
ranLevels = list(rio = rioL, mes = mesL),
distr = "lognormal poisson")Definimos los parámetros para las Cadenas de Markov Monte Carlo
Se calibran y estiman los parámetros del modelos con las MCMC
m0 <- sampleMcmc(m0,
thin = thin,
samples = samples,
transient = transient,
nChains = nChains,
nParallel = nChains)
m1 <- sampleMcmc(m1,
thin = thin,
samples = samples,
transient = transient,
nChains = nChains,
nParallel = nChains)
m_env <- sampleMcmc(m_env,
thin = thin,
samples = samples,
transient = transient,
nChains = nChains,
nParallel = nChains)
m_env_phy <- sampleMcmc(m_env_phy,
thin = thin,
samples = samples,
transient = transient,
nChains = nChains,
nParallel = nChains)Convertimos los resultados en objetos CODA, son un tipo de datos de análisis bayesianos en R
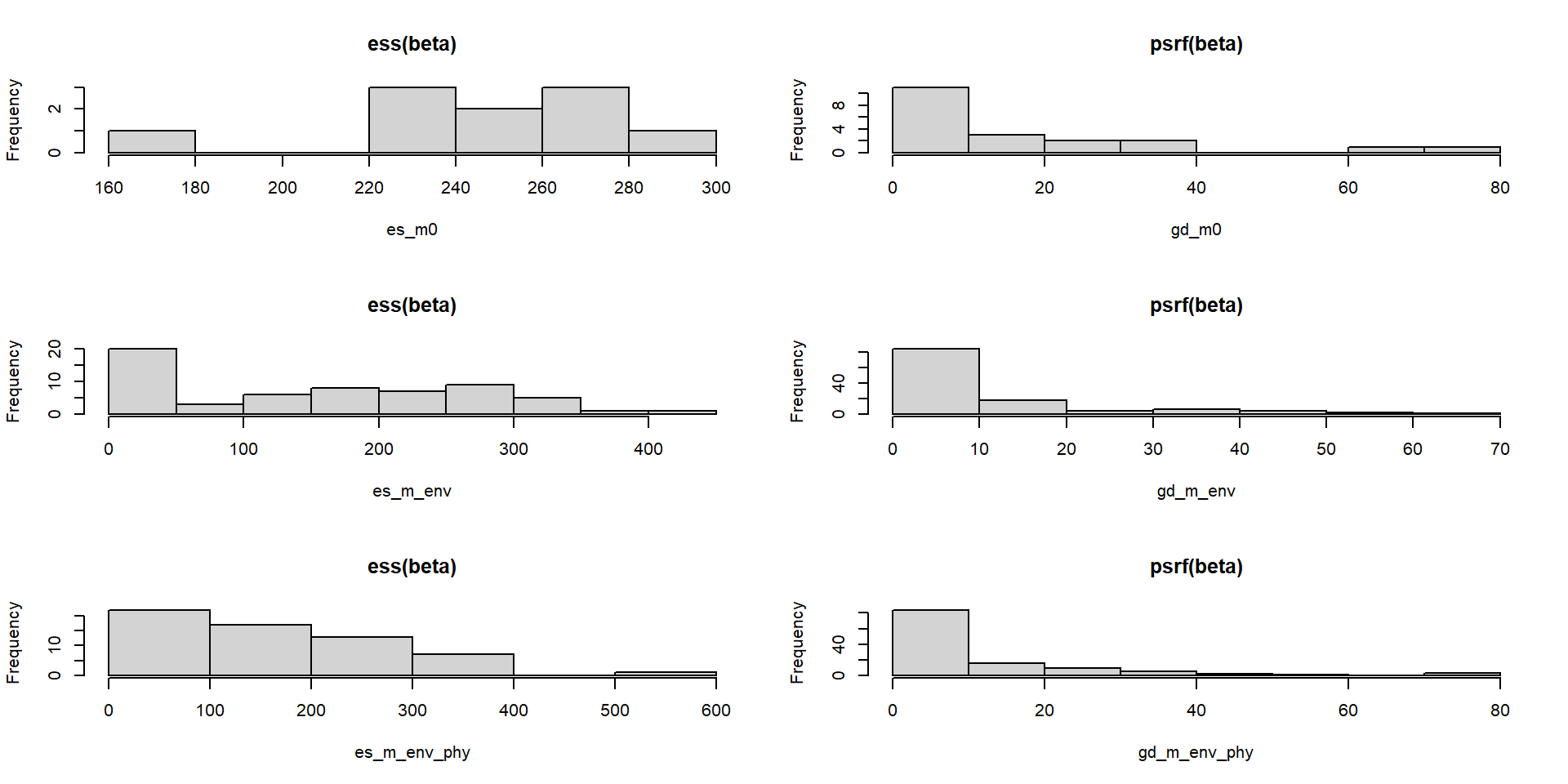
Calculamos dos estadísticos de calibración de MCMC
Dibujamos las gráficas de esos dos parámetros
Effective size debería estar entorno al número de muestras de las MCMC (samples * nChains). El diagnóstico de Gelman debería estar entorno a 1.
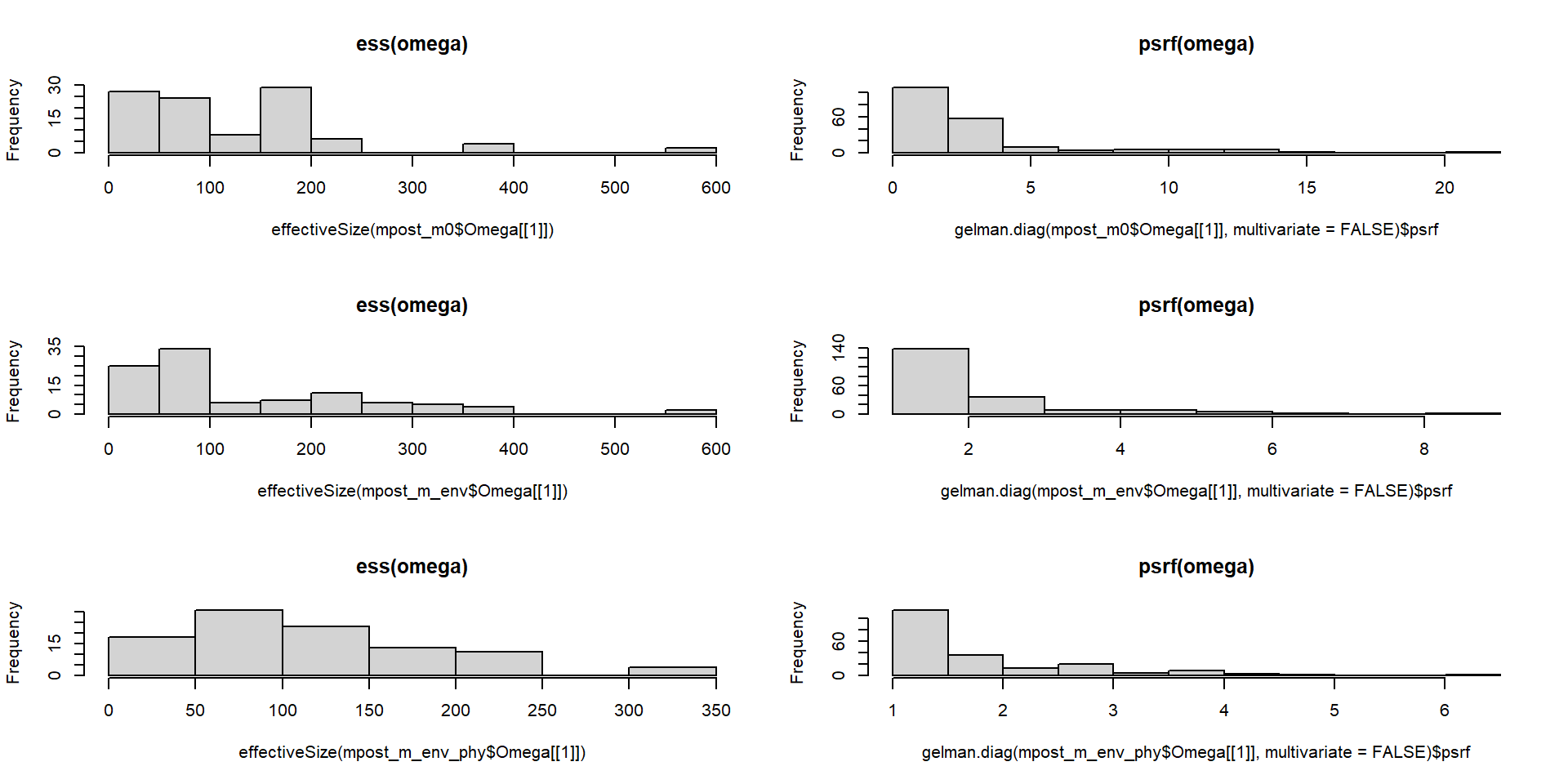
… lo mismo pero con los valores de Omega, que son los coeficientes de interacciones entre especies
par(mfrow=c(3,2))
hist(effectiveSize(mpost_m0$Omega[[1]]), main="ess(omega)")
hist(gelman.diag(mpost_m0$Omega[[1]], multivariate=FALSE)$psrf, main="psrf(omega)")
hist(effectiveSize(mpost_m_env$Omega[[1]]), main="ess(omega)")
hist(gelman.diag(mpost_m_env$Omega[[1]], multivariate=FALSE)$psrf, main="psrf(omega)")
hist(effectiveSize(mpost_m_env_phy$Omega[[1]]), main="ess(omega)")
hist(gelman.diag(mpost_m_env_phy$Omega[[1]], multivariate=FALSE)$psrf, main="psrf(omega)")Tambien podemos dibujar algunas de las cadenas para visulizar su forma y ver si se han estabilizado
Para evaluar el modelo en términos de la distribución/abundancia de las especies, debemos hacer predicciones primero.
$RMSE
[1] 178.7046818 112.1060668 20.0050140 25.8061127 1.4742253 1.2257749
[7] 0.3629371 0.5948718 0.4136221 0.7108976
$SR2
[1] 0.44104705 0.43882516 0.14058017 0.33967948 0.00404370 0.12155108
[7] -0.01259728 0.05565401 0.03797939 0.06015246
$O.AUC
[1] 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5
$O.TjurR2
[1] 0 0 0 0 0 0 0 0 0 0
$O.RMSE
[1] 0.5814019 0.5566499 0.8960005 0.8049145 0.9115843 0.9786452 0.9858149
[8] 0.9786452 0.9786452 0.9858149
$C.SR2
[1] 0.3439112993 0.3598536094 0.0257322387 0.1150386032 0.0001164009
[6] NA NA NA NA NA
$C.RMSE
[1] 217.0444042 134.5360937 44.7811536 42.4439580 3.4896220 5.6977254
[7] 0.6537211 0.3809210 0.5918066 0.2427098$RMSE
[1] 239.5894479 124.3036980 19.9331009 28.0490736 1.4336509 1.2093620
[7] 0.2752305 0.3071712 0.3476443 0.2666874
$SR2
[1] 0.2094872098 0.2722988542 0.0342582931 0.1587915733 -0.0036198349
[6] 0.0643153798 0.0001552795 -0.0078898226 0.0215802988 0.0038819876
$O.AUC
[1] 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5
$O.TjurR2
[1] 0 0 0 0 0 0 0 0 0 0
$O.RMSE
[1] 0.5814019 0.5566499 0.8960005 0.8049145 0.9115843 0.9786452 0.9858149
[8] 0.9786452 0.9786452 0.9858149
$C.SR2
[1] 0.16373436 0.14570572 0.07679719 0.04803554 0.27188927 0.25000000
[7] NA NA NA NA
$C.RMSE
[1] 287.3041422 146.8263550 43.3889397 39.8621564 3.3175464 5.7124472
[7] 0.8489667 0.8425653 0.6761803 0.7612070$RMSE
[1] 198.0809861 121.5655796 19.7023785 25.8081348 1.4474939 1.1704319
[7] 0.3259737 0.2917941 0.2623690 0.2711962
$SR2
[1] 0.3113252526 0.4452635687 0.0845909888 0.2838581631 -0.0005258901
[6] 0.0908541598 -0.0008454106 0.0042133520 0.0280228758 0.0033816425
$O.AUC
[1] 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5
$O.TjurR2
[1] 0 0 0 0 0 0 0 0 0 0
$O.RMSE
[1] 0.5814019 0.5566499 0.8960005 0.8049145 0.9115843 0.9786452 0.9858149
[8] 0.9786452 0.9786452 0.9858149
$C.SR2
[1] 0.24034333 0.27846395 0.07058145 0.12653319 0.26079293 1.00000000
[7] NA NA NA NA
$C.RMSE
[1] 242.4261169 145.5229074 43.8888197 40.7282525 3.4359286 5.5406217
[7] 0.7611457 0.8769854 0.7229513 0.7906554preds_m_env_phy <- computePredictedValues(m_env_phy)
evaluateModelFit(hM = m_env_phy, predY = preds_m_env_phy)$RMSE
[1] 216.0099471 121.4572935 19.4606626 25.7175698 1.4555382 1.2045312
[7] 0.2779336 0.4405696 0.7282538 0.2475604
$SR2
[1] 3.682232e-01 4.713293e-01 1.199537e-01 3.359516e-01 2.578516e-02
[6] 9.073820e-02 -1.552795e-04 2.689076e-02 1.685341e-02 6.901311e-05
$O.AUC
[1] 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5
$O.TjurR2
[1] 0 0 0 0 0 0 0 0 0 0
$O.RMSE
[1] 0.5814019 0.5566499 0.8960005 0.8049145 0.9115843 0.9786452 0.9858149
[8] 0.9786452 0.9786452 0.9858149
$C.SR2
[1] 0.25446799 0.30267563 0.17566157 0.07310235 0.16254407 1.00000000
[7] NA NA NA NA
$C.RMSE
[1] 264.5851587 145.7033563 43.2717989 41.7909492 3.4773240 5.6963881
[7] 0.7905187 0.6136172 0.4047374 0.7921033También es posible realizar evaluación por validación cruzada
partition_env <- createPartition(m_env, nfolds = 4)
preds_m_env = computePredictedValues(m_env,
partition = partition_env,
nParallel = nChains)Cross-validation, fold 1 out of 4
Cross-validation, fold 2 out of 4
Cross-validation, fold 3 out of 4
Cross-validation, fold 4 out of 4$RMSE
[1] 228.5461386 166.9150113 20.1004411 28.3106774 1.4873049 1.2555333
[7] 0.2856651 0.3612464 0.3271817 0.2471069
$SR2
[1] 1.205130e-01 2.006534e-01 1.955293e-02 4.666595e-02 -1.003100e-02
[6] 1.305100e-03 6.901311e-05 -2.987862e-03 1.512605e-02 -5.590062e-03
$O.AUC
[1] 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5
$O.TjurR2
[1] 0 0 0 0 0 0 0 0 0 0
$O.RMSE
[1] 0.5814019 0.5566499 0.8960005 0.8049145 0.9115843 0.9786452 0.9858149
[8] 0.9786452 0.9786452 0.9858149
$C.SR2
[1] 0.074497388 0.096339360 0.004112355 0.001445167 -0.008351153
[6] -0.250000000 NA NA NA NA
$C.RMSE
[1] 278.9709740 198.4772048 44.9055812 43.1712286 3.5037181 5.8779952
[7] 0.8099910 0.8245285 0.6666862 0.8732730Una vez que tengamos un modelo sólido. Nos interesa ver que papel juegan las variables ambientales en las especies/comunidades.
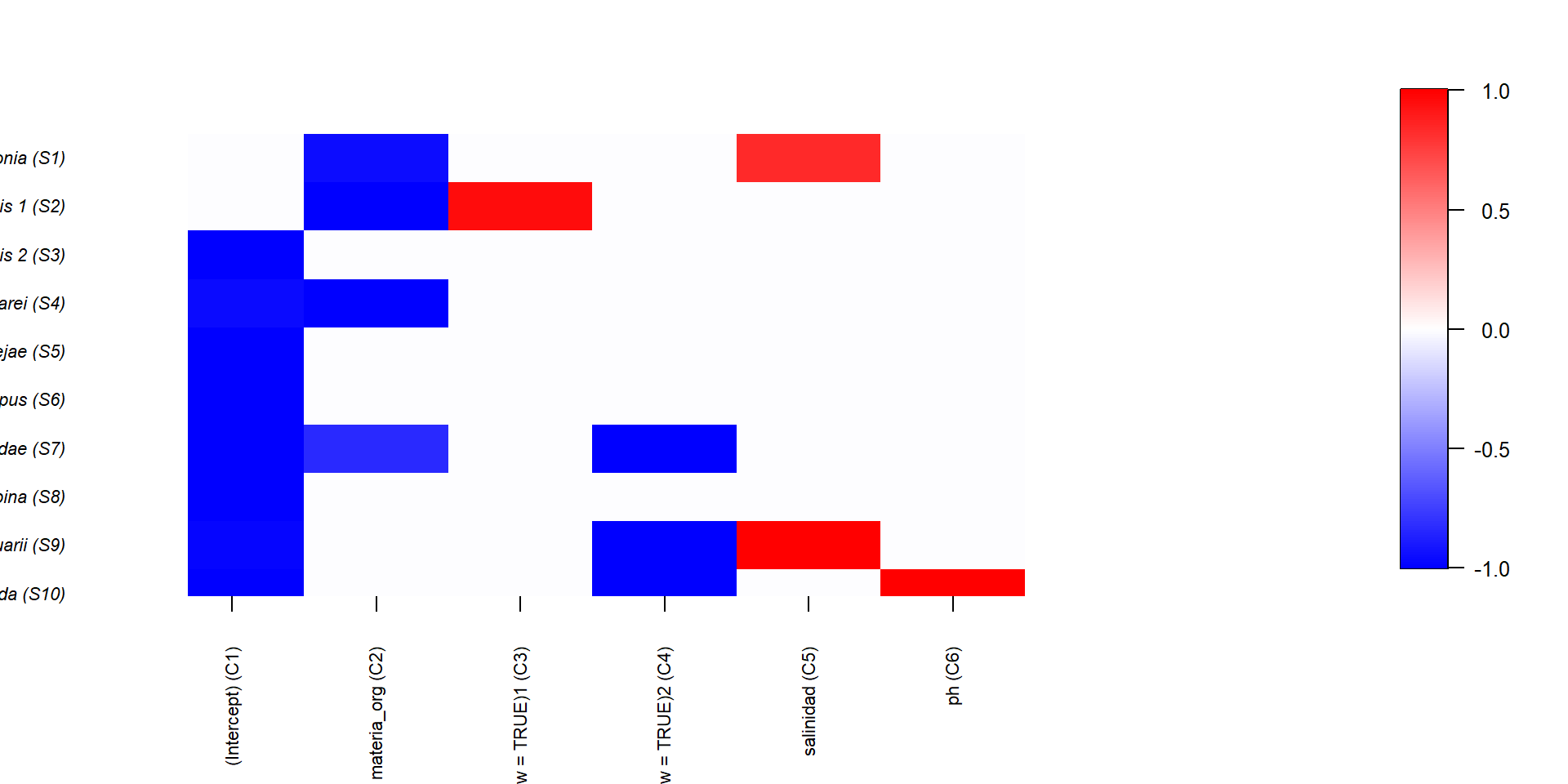
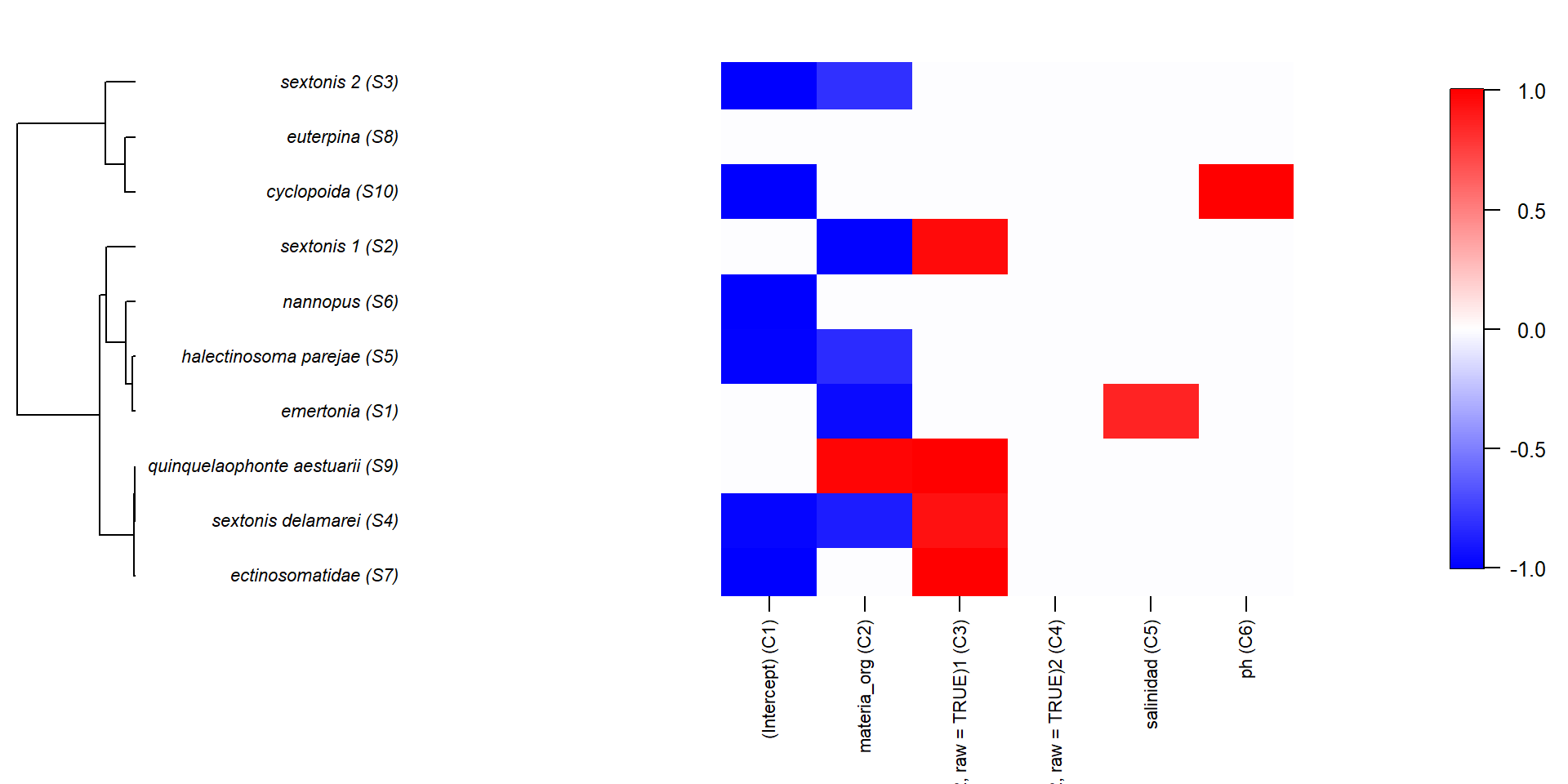
También podemos ver su relación con la información filogenética, si la hemos incluido en el modelo
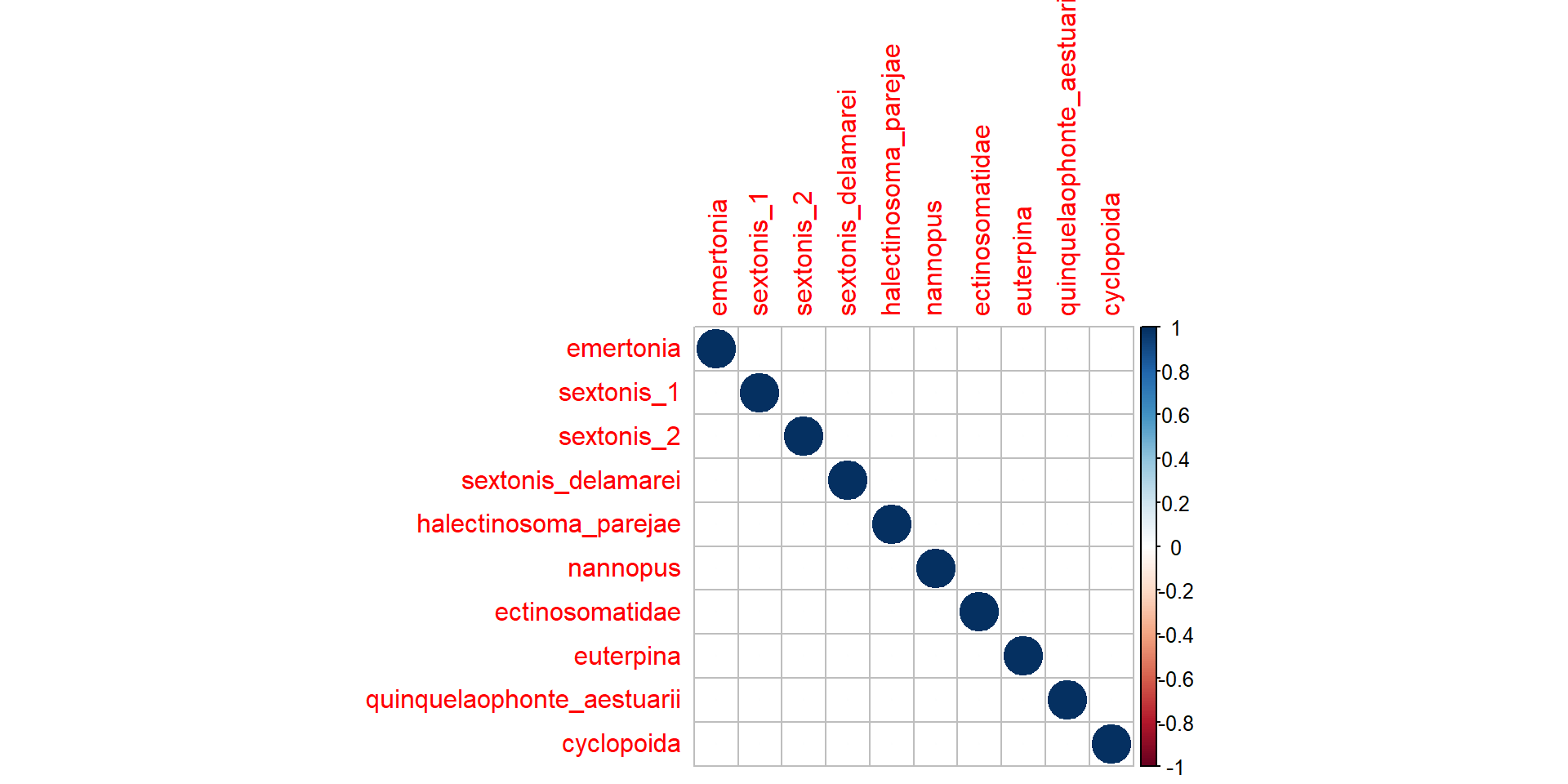
Otro aspecto importante/novedoso es estimar las correlaciones entre especies
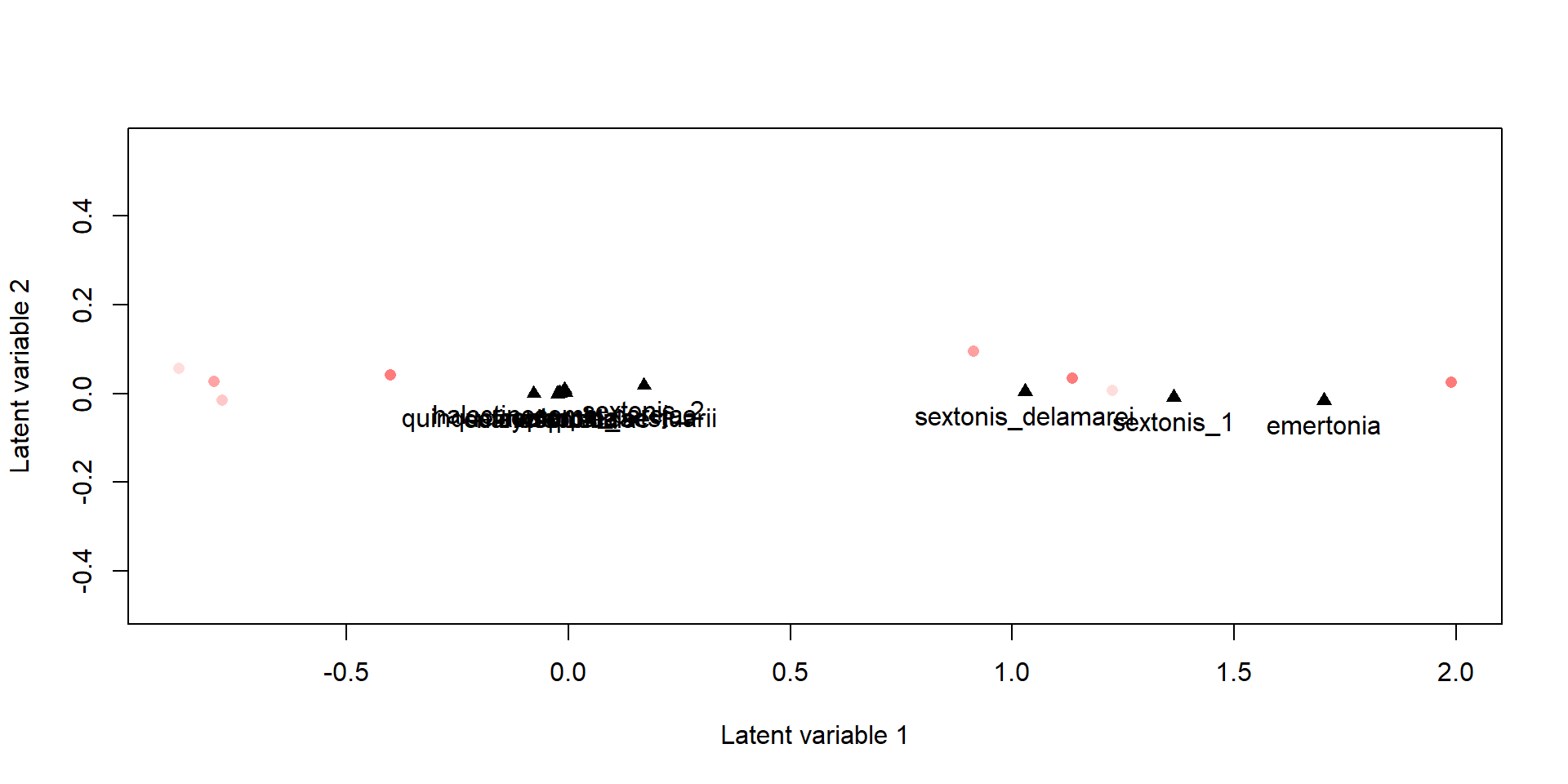
Cuando especificamos un modelo sin variables ambientales, estamos haciendo un modelo “unconstrained” o indirecto. Por lo que se puede usar para generar gráficos de ordenación.
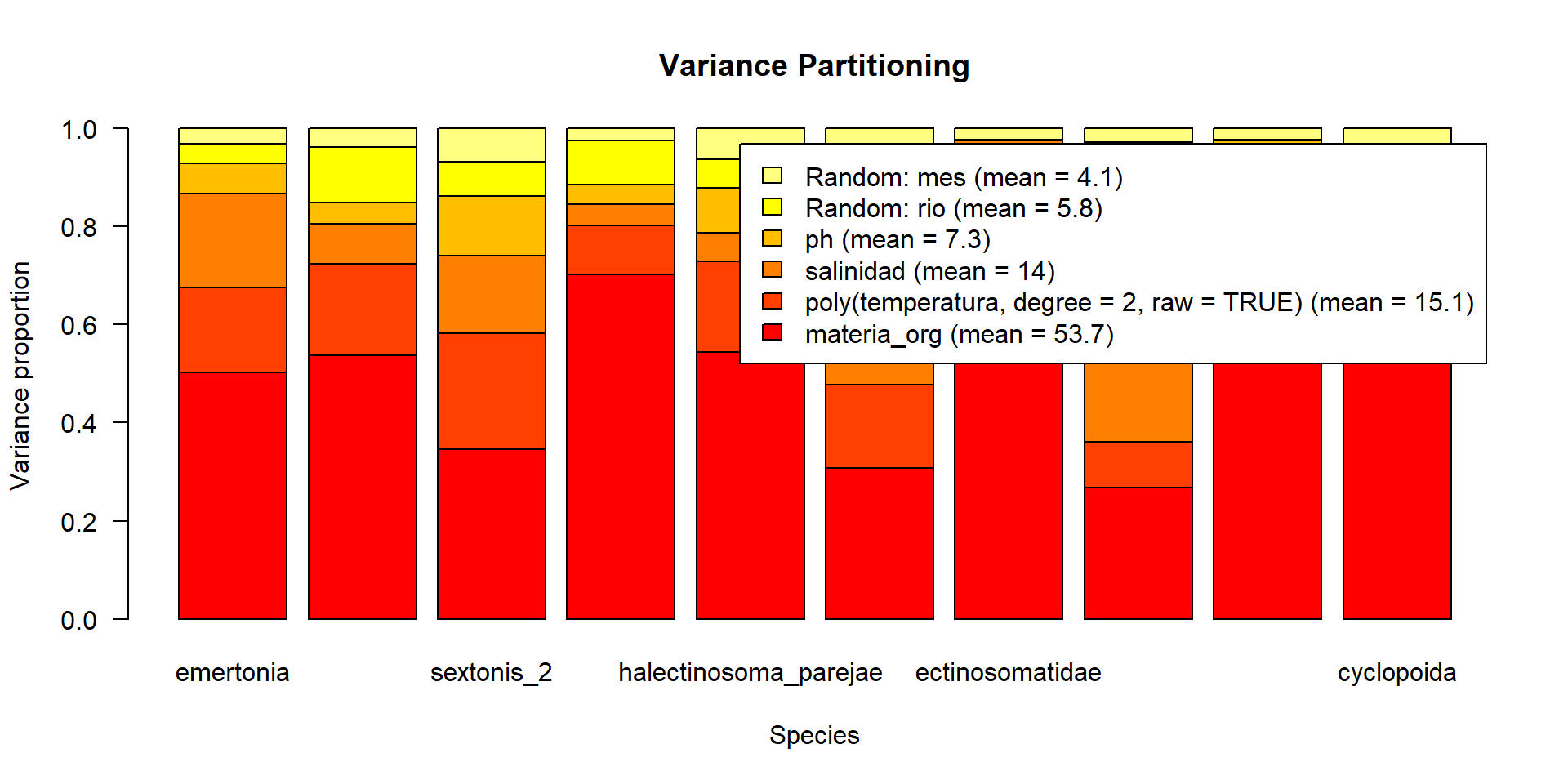
En este tipo de modelos, la importancia de cada variable se mide en términos de variación explicada y se calcula con análisis de particionado de la varianza
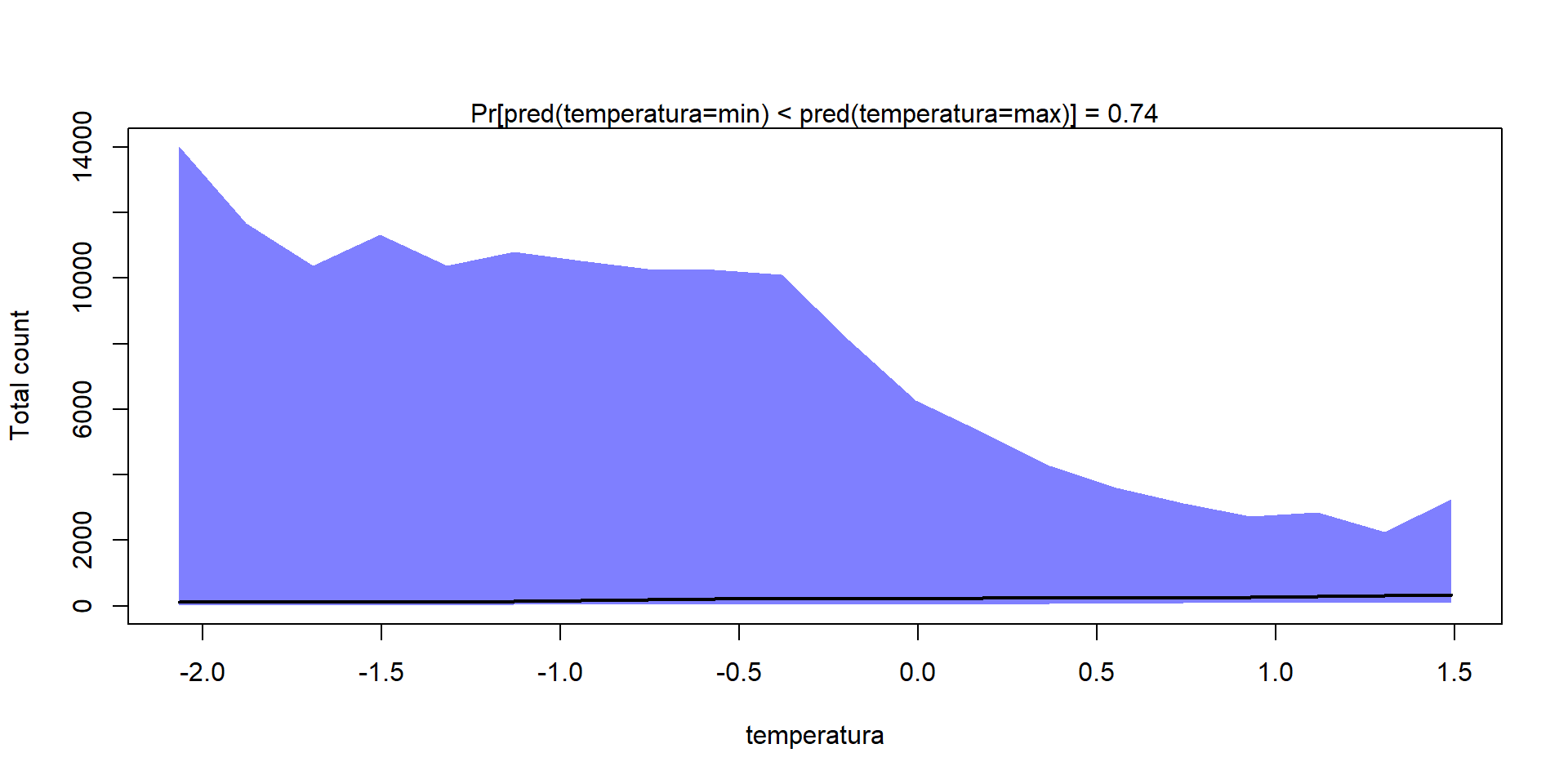
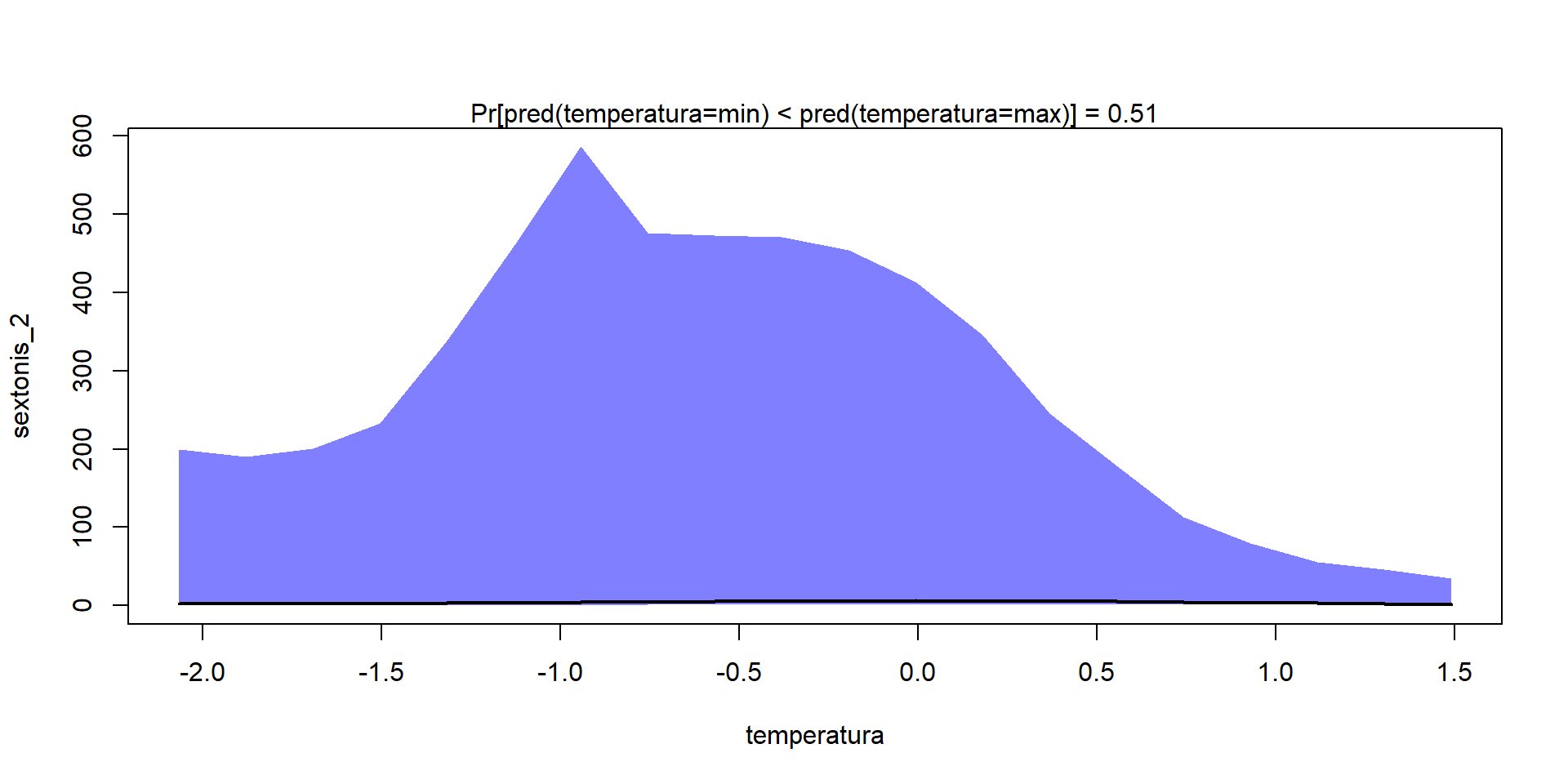
Como en los Modelos de Distribución de Especies, podemos calcular curvas de respuesta de cada especie/comunidad a las variables ambientales
Gradient = constructGradient(m1, focalVariable = "temperatura",
non.focalVariables = list("habitat"=list(3,"open")))
Gradient$XDataNew temperatura materia_org salinidad ph
1 -2.065315281 -0.1667234554 -0.501380262 0.37285247
2 -1.878146837 -0.1516142030 -0.455942859 0.33906285
3 -1.690978393 -0.1365049507 -0.410505456 0.30527323
4 -1.503809949 -0.1213956984 -0.365068052 0.27148361
5 -1.316641505 -0.1062864460 -0.319630649 0.23769400
6 -1.129473060 -0.0911771937 -0.274193245 0.20390438
7 -0.942304616 -0.0760679413 -0.228755842 0.17011476
8 -0.755136172 -0.0609586890 -0.183318438 0.13632514
9 -0.567967728 -0.0458494366 -0.137881035 0.10253552
10 -0.380799284 -0.0307401843 -0.092443632 0.06874590
11 -0.193630840 -0.0156309320 -0.047006228 0.03495628
12 -0.006462395 -0.0005216796 -0.001568825 0.00116666
13 0.180706049 0.0145875727 0.043868579 -0.03262296
14 0.367874493 0.0296968251 0.089305982 -0.06641258
15 0.555042937 0.0448060774 0.134743386 -0.10020220
16 0.742211381 0.0599153298 0.180180789 -0.13399182
17 0.929379825 0.0750245821 0.225618193 -0.16778144
18 1.116548270 0.0901338344 0.271055596 -0.20157106
19 1.303716714 0.1052430868 0.316492999 -0.23536068
20 1.490885158 0.1203523391 0.361930403 -0.26915030predY <- predict(m1,
XData=Gradient$XDataNew,
studyDesign=Gradient$studyDesignNew,
ranLevels=Gradient$rLNew,
expected=TRUE)
plotGradient(m1, Gradient, pred=predY, measure="S")[1] 0.74[1] 0.5125